Demo#
A poisson regression is used to model counts and/or rates. A count is zero or any positive integer, such as the number of events, the number of students, etc.
A poisson regression (log-linear model) uses a log link function to relate the linear component (\(\eta\)) to the \((0,\infty)\) count variable, e.g. (0,1,2…)
To model rates, we include an “offset” term:
Then Y follows a Poisson distribution parameterized by \(\mu\):
import numpy as np
import pandas as pd
import statsmodels.api as sm
import seaborn as sns
import patsy
sns.set(rc={'figure.figsize':(11.7,8.27)})
Generate Fake Data#
We generate two independent variables x1 and x2. We let \(\eta\) be the linear component, \(X\beta\).
To get \(\mu\), we exponentiate both sides of \(log(\mu) = \eta\), so we have \(\mu = exp(\eta)\)
Then y (# of students flagged) is poisson distributed, with its single parameter: \(\mu\).
To demonstrate poisson modeling with rates, we create an offset term: n_enrolled, which is simply n_student_flagged plus a random positive integer.
n = 500
np.random.seed(0)
df = pd.DataFrame({
"x1":np.random.exponential(2,n),
"x2":np.random.normal(2,1,n),
})
eta = np.exp(2 + 0.25*df["x1"] + 0.25*df["x2"])
df["n_students_flagged"] = np.random.poisson(lam=eta)
df["n_students_flagged"].value_counts()
df["n_enrolled"] = np.round(df["n_students_flagged"],0) + np.random.binomial(200,0.8,n)
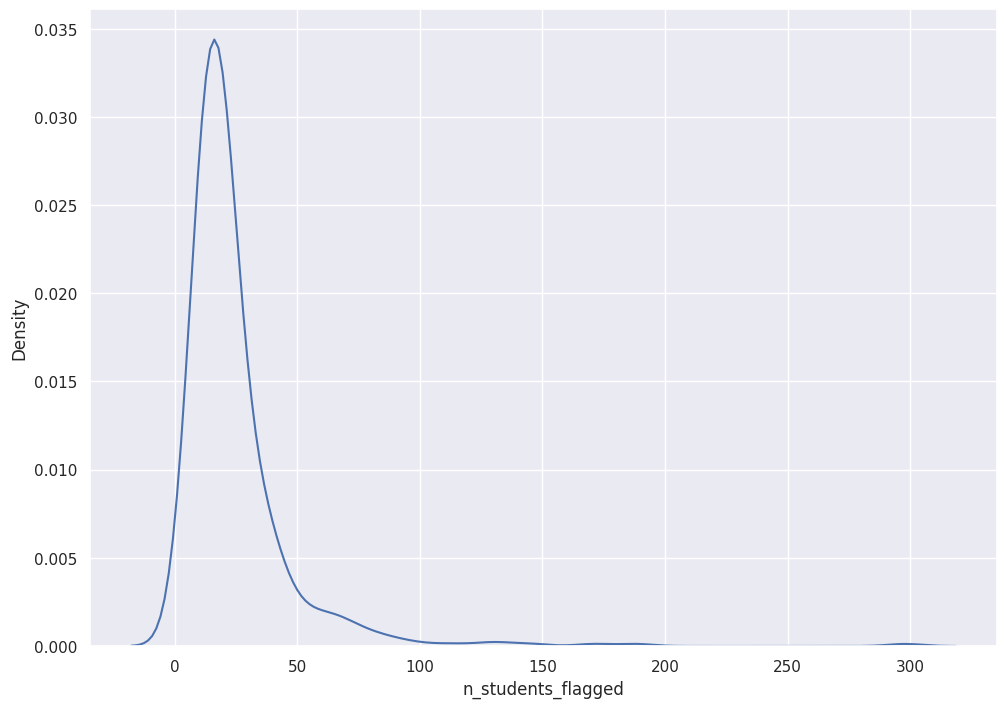
sns.kdeplot(df["n_students_flagged"])
<Axes: xlabel='n_students_flagged', ylabel='Density'>
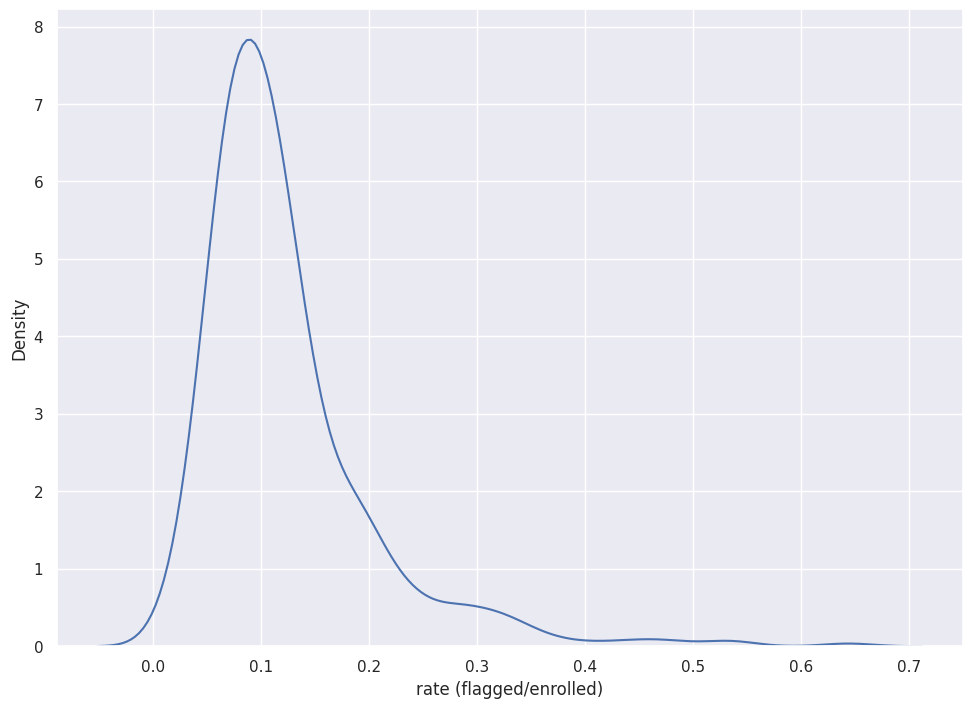
g = sns.kdeplot(df["n_students_flagged"]/df["n_enrolled"])
g.set_xlabel("rate (flagged/enrolled)")
Text(0.5, 0, 'rate (flagged/enrolled)')
f = 'n_students_flagged ~ x1 + x2'
y, X = patsy.dmatrices(f, df, return_type='dataframe')
Statsmodels#
mod = sm.GLM(y, X, family=sm.families.Poisson())
res2 = mod.fit()
print(res2.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: n_students_flagged No. Observations: 500
Model: GLM Df Residuals: 497
Model Family: Poisson Df Model: 2
Link Function: Log Scale: 1.0000
Method: IRLS Log-Likelihood: -1486.9
Date: Thu, 01 Jun 2023 Deviance: 571.92
Time: 12:45:56 Pearson chi2: 563.
No. Iterations: 5 Pseudo R-squ. (CS): 1.000
Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 2.0270 0.025 79.737 0.000 1.977 2.077
x1 0.2469 0.003 86.086 0.000 0.241 0.253
x2 0.2447 0.010 25.097 0.000 0.226 0.264
==============================================================================
using n_enrolled as offset to model rate (flagged out of enrolled) instead of count (# enrolled)
including an offset is equivalent to including the offset as another pedictor, except with the coefficient fixed to 1.
mod = sm.GLM(y, X, offset=df["n_enrolled"], family=sm.families.Poisson())
res2 = mod.fit()
print(res2.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: n_students_flagged No. Observations: 500
Model: GLM Df Residuals: 497
Model Family: Poisson Df Model: 2
Link Function: Log Scale: 1.0000
Method: IRLS Log-Likelihood: -2.5985e+35
Date: Thu, 01 Jun 2023 Deviance: 5.1970e+35
Time: 12:45:56 Pearson chi2: 2.68e+37
No. Iterations: 100 Pseudo R-squ. (CS): nan
Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
Intercept -99.7842 2.52e-18 -3.97e+19 0.000 -99.784 -99.784
x1 -24.1414 2.02e-17 -1.19e+18 0.000 -24.141 -24.141
x2 -2.5238 4.29e-18 -5.88e+17 0.000 -2.524 -2.524
==============================================================================
/home/chansoo/projects/statsbook/.venv/lib/python3.8/site-packages/statsmodels/genmod/families/family.py:132: RuntimeWarning: divide by zero encountered in divide
return 1. / (self.link.deriv(mu)**2 * self.variance(mu))