Applied#
from sklearn.cluster import AgglomerativeClustering
from scipy.cluster.hierarchy import dendrogram
from sklearn.datasets import make_blobs
import matplotlib.pyplot as plt
plt.figure(figsize=(16, 14), dpi=80)
import numpy as np
centers = [(-3, -3), (4, 4), (4, -4)]
cluster_std = [2, 3, 2]
X, y = make_blobs(
n_samples=500,
cluster_std = cluster_std,
centers = centers,
n_features = 2,
random_state=0
)
# https://scikit-learn.org/stable/auto_examples/cluster/plot_agglomerative_dendrogram.html#sphx-glr-auto-examples-cluster-plot-agglomerative-dendrogram-py
def plot_dendrogram(model, **kwargs):
# Create linkage matrix and then plot the dendrogram
# create the counts of samples under each node
counts = np.zeros(model.children_.shape[0])
n_samples = len(model.labels_)
for i, merge in enumerate(model.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
plt.figure()
dendrogram(linkage_matrix, **kwargs)
<Figure size 1280x1120 with 0 Axes>
Using sklearn#
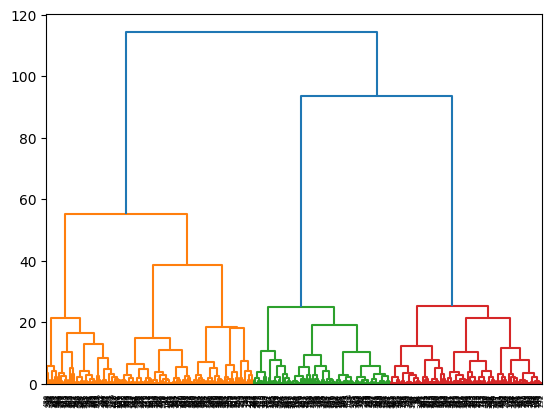
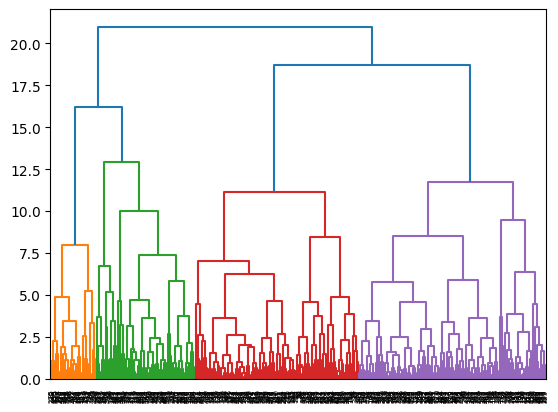
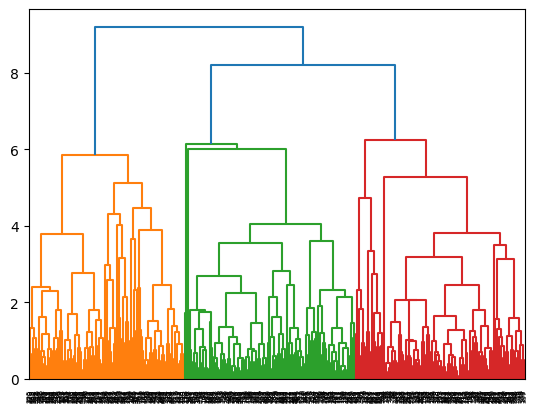
for i,linkage in enumerate(['ward', 'complete', 'average']):
cluster = AgglomerativeClustering(
linkage=linkage,
distance_threshold=0,
n_clusters=None
)
cluster = cluster.fit(X)
plot_dendrogram(cluster)